Opportunity for AI
It is well known that children get sick more often, as part of the natural process of developing their own immune systems. Consequently, the need for diagnostics is much more frequent, putting the burden on parents. According to data from the National Health Interview Survey, in 2019, more than one in four children had one or more visits to an urgent care center or retail health clinic (26.4%) in the past 12 months.
Unnecessary visits to ER represent an enormous waste of valuable medical resources, not to mention the economical and societal cost, as well as impact on the well-being of each individual family. According to a multi-year analysis of children’s visit to ER in hospitals in Italy, 75.8% of the visits are unnecessary. Most of the ER visits are result from independent decision of the parents (97.2%), especially in the evening and at night on Saturdays/Sundays/holidays (69.7%).
A distinguishing characteristic with children’s diagnostics is the high percentage of cases resulting from common illness. In the above study, the most common trigger resulting in parents’ decision to visit ER was fever (51.4%)
Opportunity is ripe for an AI system which can identify the most common illness that does not require ER visits with a certain degree of accuracy. There is no need to provide diagnostics for complex diseases. That is a fundamental consideration when it comes to collecting data, designing, and building such a system.
Proposed Design
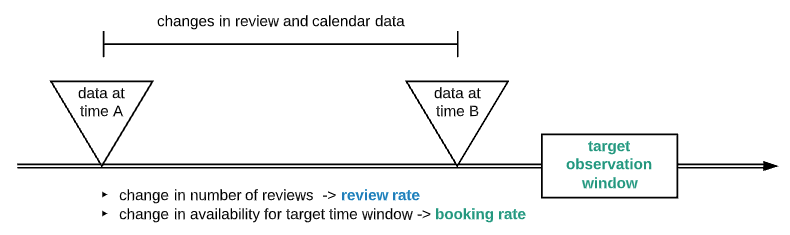
Illustrated below is a proposed design which is based on the following principles:
- Simulating the knowledge and iterative diagnostic process of a physician
- Utilizing combination of AI technologies including NLP, vision models
- Focusing on well-defined target outcome (diagnostics of common childhood illness)
Here are highlights of how the system works:
- Allow user to start with free text description of the symptoms and illness
- With a predefined list of symptoms, use transformer model to do Reverse Asymmetric Semantic Search against the user query, resulting in a list of matching symptoms (binary features)
- With well defined symptom-disease data (see next section), use a classification model to predict a list of candidate diseases based on symptoms
- Based on the candidates identify additional information to probe user for, iteratively predict/probe until threshold is met
- Incorporate additional model into the system (image, video)
- Arrive at diagnostic (or no diagnostic), present result and recommendation to the user
Prototype - Proof of Concept
Illustrated below is a prototype built to demonstrate the concept and how different AI components work together to form a more sophisticated system.
The prototype is built around two machine learning models, an NLP model that is designed to turn free text input into symptom features. The symptom features are fed into a symptom-disease predictor to get a diagnostic.
More details are provided in the following sections.
Sample Disease/symptoms Data
Below shows the sample dataset used, each patient case is diagnosed with a disease, together with observed symptoms.
To prepare data for training, we encode each patient case, with disease as the prediction target, and symptoms as encoded features.
Predictor model
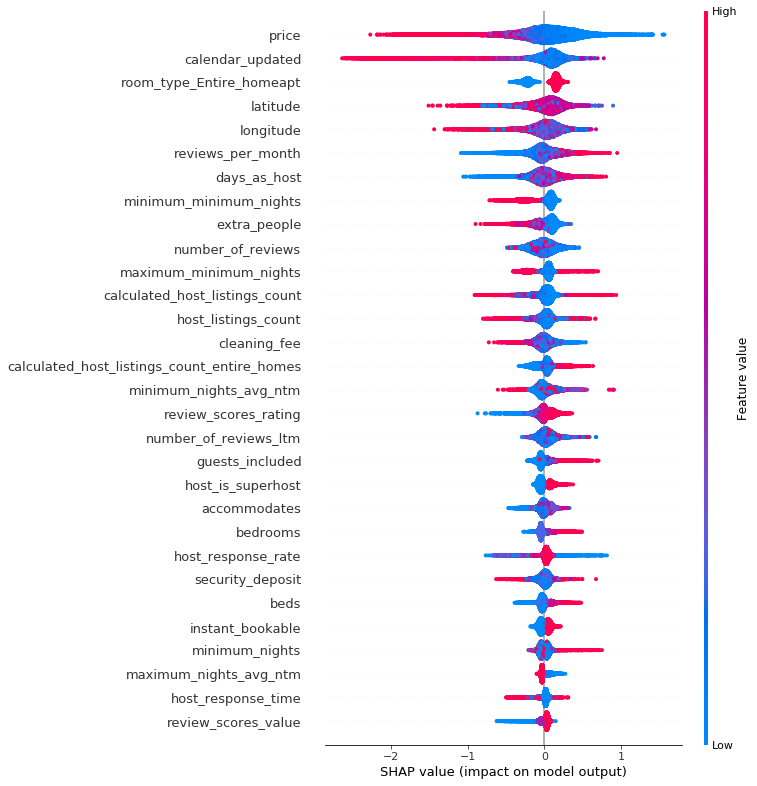
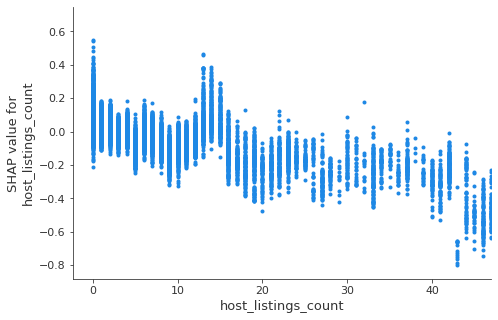
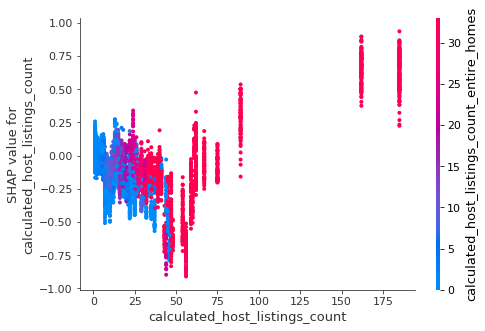
We can use gradient boosting to train a classification model that predicts disease based on symptoms. Here is the outcome of a LightGBM model, showing both the overall accuracy and per disease.
User Query
With this component, we use a transformer model to vectorize all the symptoms, and the incoming NLP query. By performing asymmetric semantic search with each symptom, we get a list of “activated” or matched symptoms.
We choose a threshold to apply against above matching score, to generate symptom feature for the query case, now we are ready to make a prediction using previously trained disease predictor model.
Use previously trained classification model to predict the disease:
Github
Sample code for prototype can be found here: https://github.com/seanxwang/pediatric_self_diagnosis